7. Gene ontology database: structure and validation of the data...
- Look at the definition of ontology in wikipedia
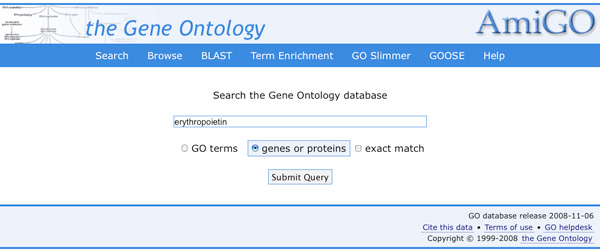
- Go to the Gene ontology consortium (AmiGO)
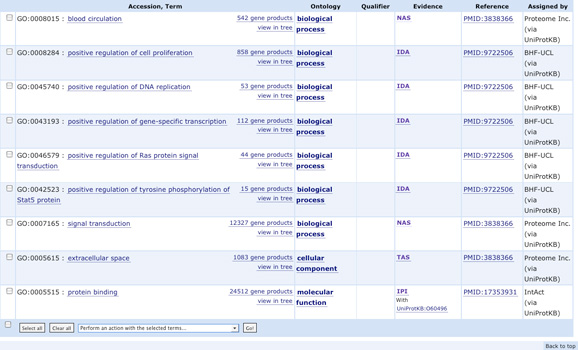
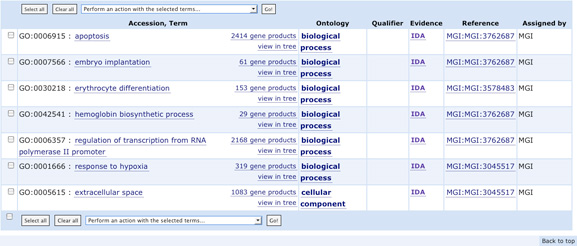
- Compare the GO terms associated with mouse and human erythropoeitin
-
Go to the UniProtKB/Swiss-Prot entry for human erythropoietin: look at
the associated GO terms How many terms have been 'manually' attributed
to the gene (have a look to the GO evidence tag )?
Remark: the GO database is of good quality.
The problem is the usage of these GO terms: they are attributed automatically to a given
gene, according to the results of protein sequence analysis (=prediction), i.e. domains
(InterPro results), subcellular location etc…