Instructions for plugin teaching
This guide is a step-by-step manual for creating and using a
training set that can be applied for the trainable calculations of Calculator
Plugins (pKa, logP).
The training of the plugin starts with a file where you collect your
experimental data (step 1, see below). From this, you generate a file which serves as knowledge
base for the plugin (step 2), that you have to copy to a directory where the plugin
picks it for usage (step 3). The plugin calculation will consider this knowledge base
file if you turn on this option when running the calculation (step 4).
Training of the pKa plugin
- Create a training set in sdfile (.sdf)
format from your experimental data. The file must
contain the following fields:
- structure
- pka value 1 (field name: pKa1)
- ID of the atom which has the pKa1 value (field name: ID1)
Additional pKa values are optional (recommended for handling multiprotic compunds):
- pKa value 2 (pKa2)
- ID2
- etc.
Definition of only one pKa value is enough to apply the
training data, but more values in case of multiprotic compounds will enhance the
reliability of the pKa teaching.
In this example this file is mydata.sdf.
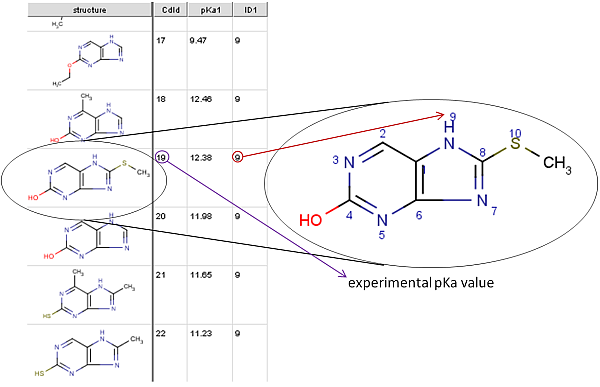
The picture below is a detail from the training file. ID1 is the index
of the atom with the experimental pKa1 value (ID2 would
be the index of the second measured pKa value /pKa2/, etc.).
This atom index can be viewed by checking the Atom number option in the molecule editor (menu: View->Misc).
- Generation of the knowledge base:
Execute the following command from command line:
cxcalc -T pka -o [path] mydata.sdf
(option -o gives the location of the folder
creates.) A 'pKaReg' folder will be created containing the training data.
Create a folder called config in the Marvin installation directory.
If Marvin's default installation was followed, Marvin installation directory is located in:
- Windows: C:\Program Files\ChemAxon\MarvinBeans
- Linux: USERHOME/ChemAxon/MarvinBeans (e.g. /home/myaccount/ChemAxon/MarvinBeans)
- OS X: /Applications/ChemAxon/MarvinBeans
- Copy the created pKaReg folder to the config folder.
- Use this knowledge base via cxcalc, Chemical Terms or Marvin. The training data helps
to calculate more accurately the pKa of the
molecules and the results are closer to the experimental values.
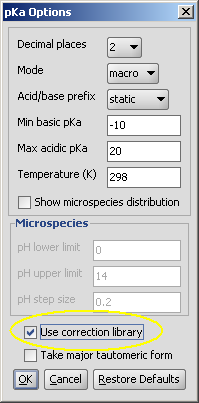
| In Marvin, check the Use correction library box to activate the training option: |
 |
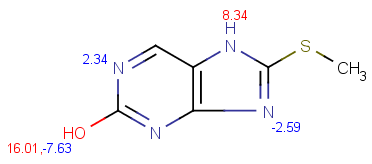
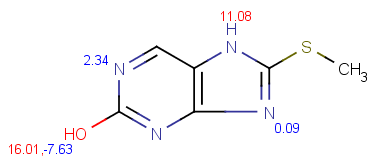
| pKa calculation without training data |
pKa calculation with training data |
|
|
- Usage with
cxcalc:
without correction library:
cxcalc pKa "CC1=NC2=C(N1)C(C)=NC(C)=N2" -a 2 -b 2
id apKa1 apKa2 bpKa1 bpKa2 atoms
1 11.08 3.67 -2.38 6,9,3
with correction library:
$ cxcalc pKa -c "CC1=NC2=C(N1)C(C)=NC(C)=N2" -a 3 -b 3
id apKa1 apKa2 bpKa1 bpKa2 atoms
1 9.90 3.67 -2.46 6,9,3
-c use the correction library
-a generate the n highest value of acidic pK
a (in this example 3)
-b generate the n highest value of basic pK
a (in this example 3)
For more options see this page.
logP training set
- Create a structure file of any molecule file format from your experimental
data. The file must contain the following information:
- structure
- logP values in a property field named LOGP
In this example this file is trainingset.sdf.
- Execute the following command from command line:
cxcalc -T logP -t LOGP -o logPparameters.txt trainingset.sdf
With the -o option you can define a path for the file generated.
Create a folder called config in the Marvin installation directory.
If Marvin's default installation was followed, Marvin installation directory is located in:
- Windows: C:\Program Files\ChemAxon\MarvinBeans
- Linux: USERHOME/ChemAxon/MarvinBeans (e.g. /home/myaccount/ChemAxon/MarvinBeans)
- OS X: /Applications/ChemAxon/MarvinBeans
- Save the file to the config folder with the name logPparameters.txt.
- Use this data via via cxcalc, Chemical Terms or Marvin.

